Xin (David) Zhao

I am a bioinformatician with a PhD in microbiology. I help life science researchers make precise and efficient discoveries by providing robust and reproducible data analysis solutions. I develop and utilize advanced computational tools, allowing scientists to concentrate on innovation and experimentation without the burden of complex data challenges.
Portfolio
Microbiome Data Analysis
Vitamin D and Infant Gut Health: A Regression Analysis
In this project, I analyzed the effects of vitamin D drops, including their non-medicinal excipients like glycerin and 1,2-propanediol, on gut metabolites and microbes in three-month-old infants. Using multiple regression models on data from the CHILD Cohort Study, I explored how vitamin D supplementation influences the infant gut environment, revealing potential impacts on gut microbiota composition and function. Learn More

Predict Childhood Obesity using RF, XGBoost and GLMM in R
Knowing obesity risk earlier could help healthcare professionals manage children’s weight more effectively. This project aims to use fecal microbiome data to predict future obesity risk with machine learning algorithms. Random Forest, XGBoost, GLMM, and Regularized Logistic classifiers were developed based on the data set of 2000+ infants. The optimal classifier achieved an AUC of 0.83 with a specificity of 73% and sensitivity of 85%. Learn More

Impact of Birth Factors on Infant Gut Health: Insights from a Microbiota Study
This academic project explores how birth factors like Caesarean sections and intrapartum antibiotic prophylaxis affect infant gut microbiota. Using linear and logistic models along with multivariate analysis, we examined the impact on Bifidobacterium abundance and overall microbiota diversity. Our findings aim to reveal how these factors shape early gut health and inform better clinical practices. Learn More

Web Application Development
WEGAN: Web Application Streamlining Ecology Data Analysis
Our collaborative project aims to create a web-based software application automating data analysis for ecologists. It utilizes the robust R package ‘vegan’ within a modern JSF framework. The user-friendly interface allows biologists to analyze their data quickly and easily, even without programming or advanced statistical expertise, generating reproducible reports in minutes.
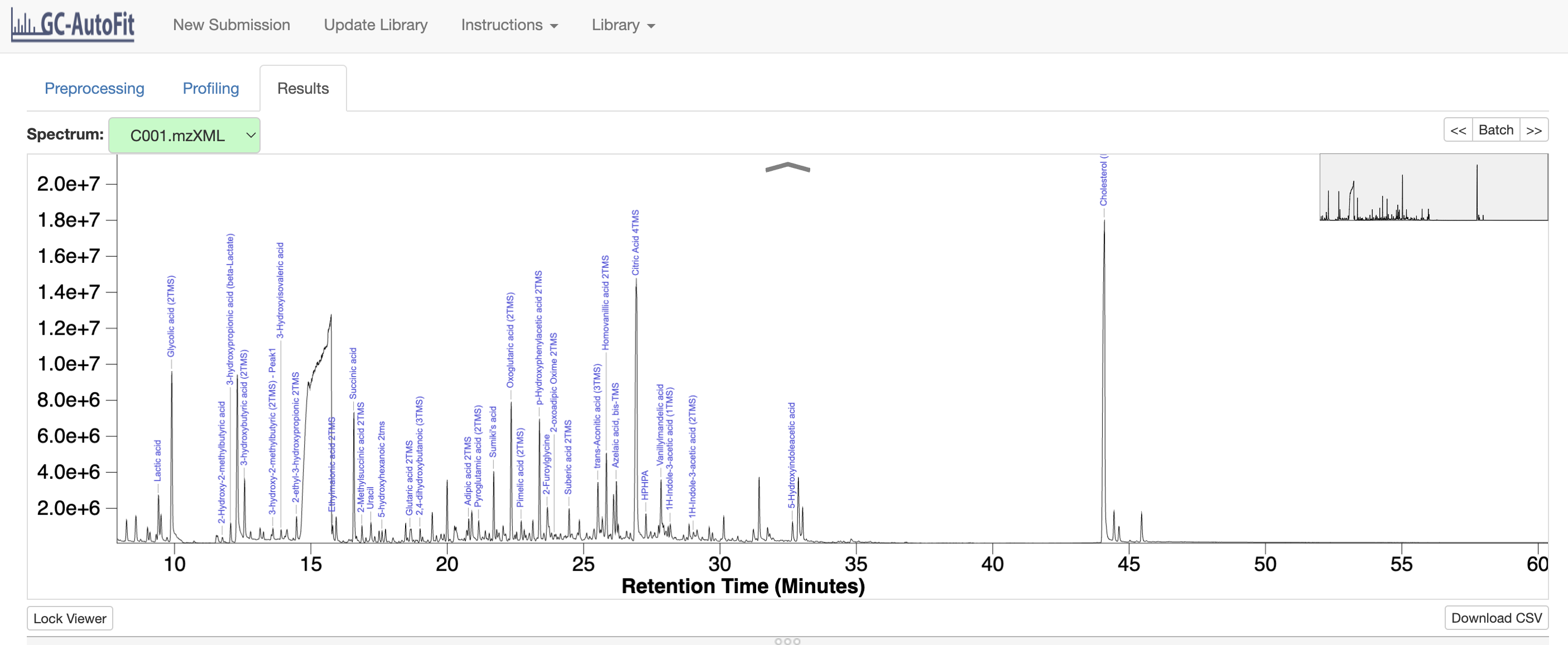
GC-AutoFit: Web Application Automating and Accelerating GC-MS Metabolomics
GC-AutoFit is a user-friendly web tool designed to make processing and analyzing data from common GC-MS systems easy, even for those without deep chemistry knowledge. It quickly identifies compounds—sometimes in just 10 seconds—and generates detailed reports that can be used with other software for further analysis. From March to August 2024, I worked on developing the first version of GC-AutoFit alongside my colleagues. I focused on building key features using Ruby on Rails for the front-end and R for the back-end, including custom inputs for biofluids and internal standards. I also debugged R scripts to ensure the accuracy of output concentrations, making GC-AutoFit both accessible and reliable for users at all levels.